 News
News
3D computer vision for brain analysis
Training AI models to identify neurons across the brain
The discovery of new ways to analyze neural data may help researchers improve the efficiency of their research into neurodegenerative disease and offer new perspectives for potential therapies.
In collaboration with the Mathis Laboratory at EPFL, we are training artificial intelligence-based computer vision models to analyze neural data from 3D microscopy datasets.
The challenge
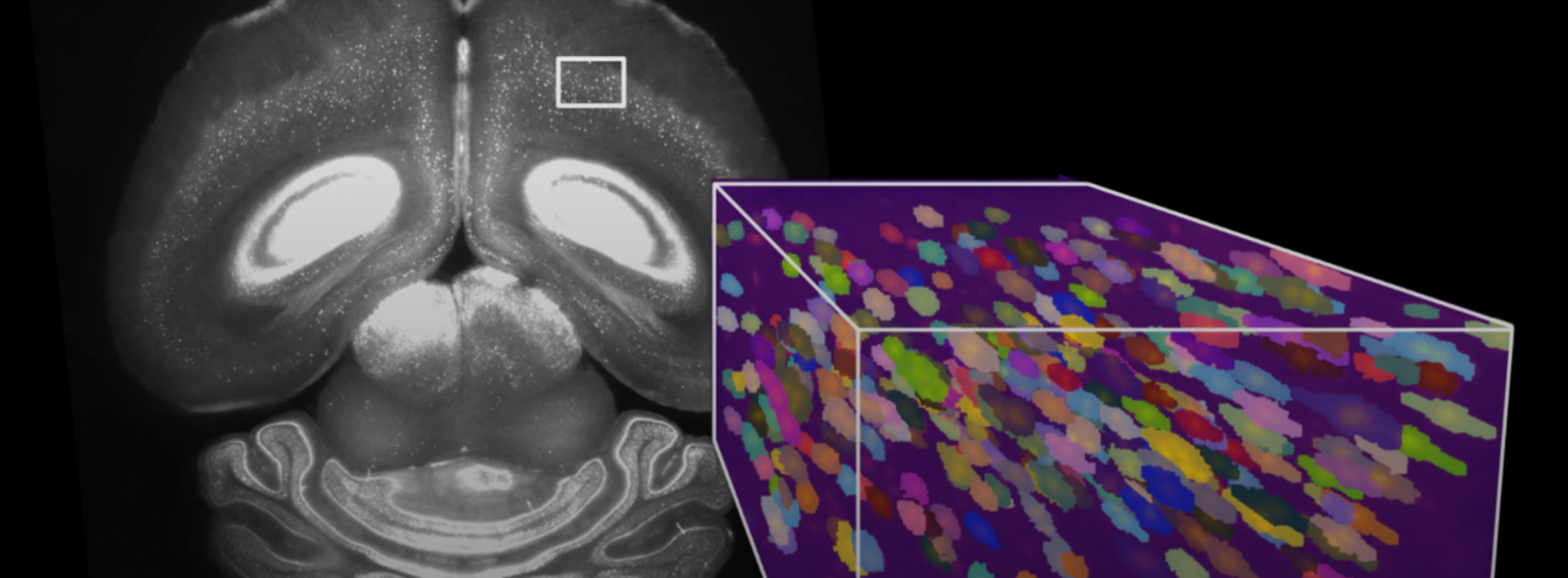
Advances in microscopy have created a petabyte-scale data avalanche and scientists are exploring the potential of computer vision to analyze the data
Currently, researchers use cell segmentation and analysis software developed for 2D data to identify and map the positions of cells in a brain, however this does not perform well with 3D data. The process is time consuming and can be inaccurate - a neuron can be mistakenly detected multiple times, or two cells can be interpreted as a single cell. Considering that the human brain has around 86 billion neurons, labelling even 0.01% of them would require the identification of millions of neurons.
At the same time, recent advances in lightsheet microscopy mean that it is now possible to image samples at subcellular spatial resolution faster than was previously possible. The new microscopes can also accommodate larger samples, from whole small brains to large sections of human brains. The high throughput of large 3D brain samples has resulted in a leap in the amount of data acquired – from megabytes to petabytes.
Now, as 3D whole-brain microscopy becomes more widespread, there is an increasing need for a rapid method of automatically and accurately detecting the multiple cell-types found within an entire brain, and the team is turning to state-of-the-art machine vision to do so.
The Mathis-Wyss Center collaboration is training computer vision models to automatically identify brain regions, accurately detect neurons and eventually track the connections between them.
MACKENZIE MATHIS, PHD, BERTARELLI FOUNDATION CHAIR OF INTEGRATIVE NEUROSCIENCE AND DIRECTOR OF THE MATHIS LABORATORY OF ADAPTIVE MOTOR CONTROL, EPFL “We are drawing on innovations in machine vision and artificial intelligence to automate the process of analyzing the millions of brain cells that we capture when we image large samples. Moreover, we also aim to provide usable tools for the broader community.”
The team is focusing on data from tissue that has been imaged with lightsheet microscopes. They are using deep neural networks to learn from large 3D data sets containing millions of brain cells.
As the project progresses, the team will expand their focus to include data from more diverse cell types involved in different brain circuits - for example dopaminergic cells involved in reward circuits and motor neurons responsible for movement - allowing them to improve the speed and accuracy of their AI models.
The resulting large data set will be open source to allow other developers to train and improve their own algorithms.
STEPHANE PAGES, PHD, SCIENTIFIC COORDINATOR AT THE WYSS CENTER AND SENIOR RESEARCH ASSOCIATE AT THE UNIVERSITY OF GENEVA “Our project is specifically geared towards developing new capabilities for microscopy image analysis of 3D brain tissue. These developments have the potential to be a real game changer in the field, allowing new discoveries and contributing to the accuracy and speed of research into neurodegenerative diseases.”
The ability to automatically detect cell type and track connections could have a major impact on the speed and reliability at which researchers learn about the mechanisms underlying human brain disorders. For example, being able to use an automated method of detecting differences in neuronal circuits would substantially increase the speed of comparing healthy and diseased brain tissue to understand the progression of neurodegenerative diseases like Alzheimer’s.